Lysogenic cycle
Lysogenic cycle also called lysogeny, is one of two cycles of viral reproduction (the lytic cycle being the other). Lysogenic cycle is characterized by integration of the bacteriophages (or phages as they are commonly known, are viruses that specifically infect bacteria) nucleic acid into the host bacterium’s genome or formation of a circular replicon in the bacterial cytoplasm. In this condition the bacterium continues to live and reproduce normally. The genetic material of the bacteriophage, called a prophage, can be transmitted to daughter cells at each subsequent cell division, and at later events (such as UV radiation or the presence of certain chemicals) can release it, causing proliferation of new phages via the lytic cycle 1. Lysogenic cycles can also occur in eukaryotes, although the method of DNA incorporation is not fully understood.
Bacteriophages or phages structure is simple, consisting of a DNA or occasionally RNA molecule carrying a number of genes, including several for replication of the phage, surrounded by a protective coat or capsid made up of protein molecules 2. In order to use a bacteriophage as a vector it is necessary to digest both target DNA and the bacteriophage DNA with restriction enzymes. Once the digestion is performed, the product will have complementary sticky ends, so both molecules can be annealed and ligated. After the resulting recombinant DNA is packaged into phage head, it can be propagated in E. coli 2.
Like all viruses, bacteriophages are very species-specific with regard to their hosts and usually only infect a single, bacterial species, or even specific strains within a species 3. Once a bacteriophage attaches to a susceptible host, it pursues one of two replication strategies: lytic or lysogenic (see Figure 1).
- In the lytic replication cycle, a phage attaches to a susceptible host bacterium, introduces its genome into the host cell cytoplasm, and utilizes the ribosomes of the host to manufacture its proteins. The host cell resources are rapidly converted to viral genomes and capsid proteins, which assemble into multiple copies of the original phage. As the host cell dies, it is either actively or passively lysed, releasing the new bacteriophage to infect another host cell.
- In the lysogenic replication cycle, the phage also attaches to a susceptible host bacterium and introduces its genome into the host cell cytoplasm. However, the phage genome is instead integrated into the bacterial cell chromosome or maintained as an episomal element where, in both cases, it is replicated and passed on to daughter bacterial cells without killing them. Integrated phage genomes are termed prophages, and the bacteria containing them are termed lysogens. Prophages can convert back to a lytic replication cycle and kill their host, most often in response to changing environmental conditions 4.
Bacteriophages are parasitic because they infect their hosts, use bacterial machinery to replicate, and ultimately lyse the bacteria. Temperate phages can lead to both advantages and disadvantages for their hosts via the lysogenic cycle. During the lysogenic cycle, the virus genome is incorporated as prophage and a repressor prevents viral replication. Nonetheless, a temperate phage can escape repression to replicate, produce viral particles, and lyse the bacteria 5. The temperate phage escaping repression would be a disadvantage for the bacteria. On the other hand, the prophage may transfer genes that enhance host virulence and resistance to the immune system. Also, the repressor produced by the prophage that prevents prophage genes from being expressed confers an immunity for the host bacteria from lytic infection by related viruses 5.
Another system, named Arbitrrium, has recently been described for bacteriophages infecting several Bacillus species, in which the decision between lysis and lysogeny is transmitted between bacteria by a peptide factor 6.
Although bacteriophages cannot infect and replicate in human cells, they are an important part of the human microbiome and a critical mediator of genetic exchange between pathogenic and non-pathogenic bacteria 7. The transfer of genes from one bacterial strain to another by a bacteriophage is called transduction and can occur in a generalized or specific manner. In “generalized” transduction, random pieces of bacterial genomic DNA are packaged inside of phage capsids in place of phage genomic DNA as the host cell is disintegrating from lytic replication. Should the phage carrying this bacterial DNA inject it into a healthy host cell, it may integrate into the chromosome of that bacterium, altering its genome and that of its daughter cells. In “specialized” transduction, it is thought that lysogenic phages, which have been amplified in a population of bacteria, excise some bacterial DNA with their genome when initiating a lytic replication cycle. Because the lysogens share the same integration site, all progeny phages transduce the same bacterial gene to their new hosts.
In addition to genetic exchange, bacteriophages can alter microbial populations because they prey on specific species of bacteria while leaving others unharmed. For more than 100 years, research has attempted to use this property as a means to treat pathogenic bacterial infections in people and animals. While wild phages probably do have transient effects on wild bacterial populations 8, many obstacles to the clinical use of lytic bacteriophages as an antimicrobial therapy (phage therapy) in humans exist. For one, wild bacterial strains are very diverse, and many are resistant to one or multiple phages. Many resistance mechanisms are known, with one famous example, the CRISPR-Cas9 system now engineered as a tool for genetic manipulation in the lab, originated as a bacterial defense mechanism against bacteriophage infection. In addition, phages are much more immunogenic than antimicrobial drugs and are rapidly cleared from the blood by the reticular endothelial system. Their large size relative to antimicrobial drugs also will likely limit their use to topical applications if effective phage cocktails are found. Some investigators have suggested that using phage enzymes which can penetrate bacterial cell walls may be a more straightforword strategy 9. To date, there have been no randomized, controlled, double-blind trials showing efficacy of either strategy in humans.
Figure 1. Life cycle of a bacteriophage
Footnote: Bacteriophages reproduce by a lytic or a lysogenic cycle. During the lytic cycle there is a phage DNA replication followed by synthesis of capsid proteins and a lysis of the bacterial cell after releasing the new phages. Instead, lysogeny is characterized by integration of the phage nucleic acid into the host bacterium’s genome. The genetic material of the phage can be transmitted to daughter cells at each subsequent cell division.
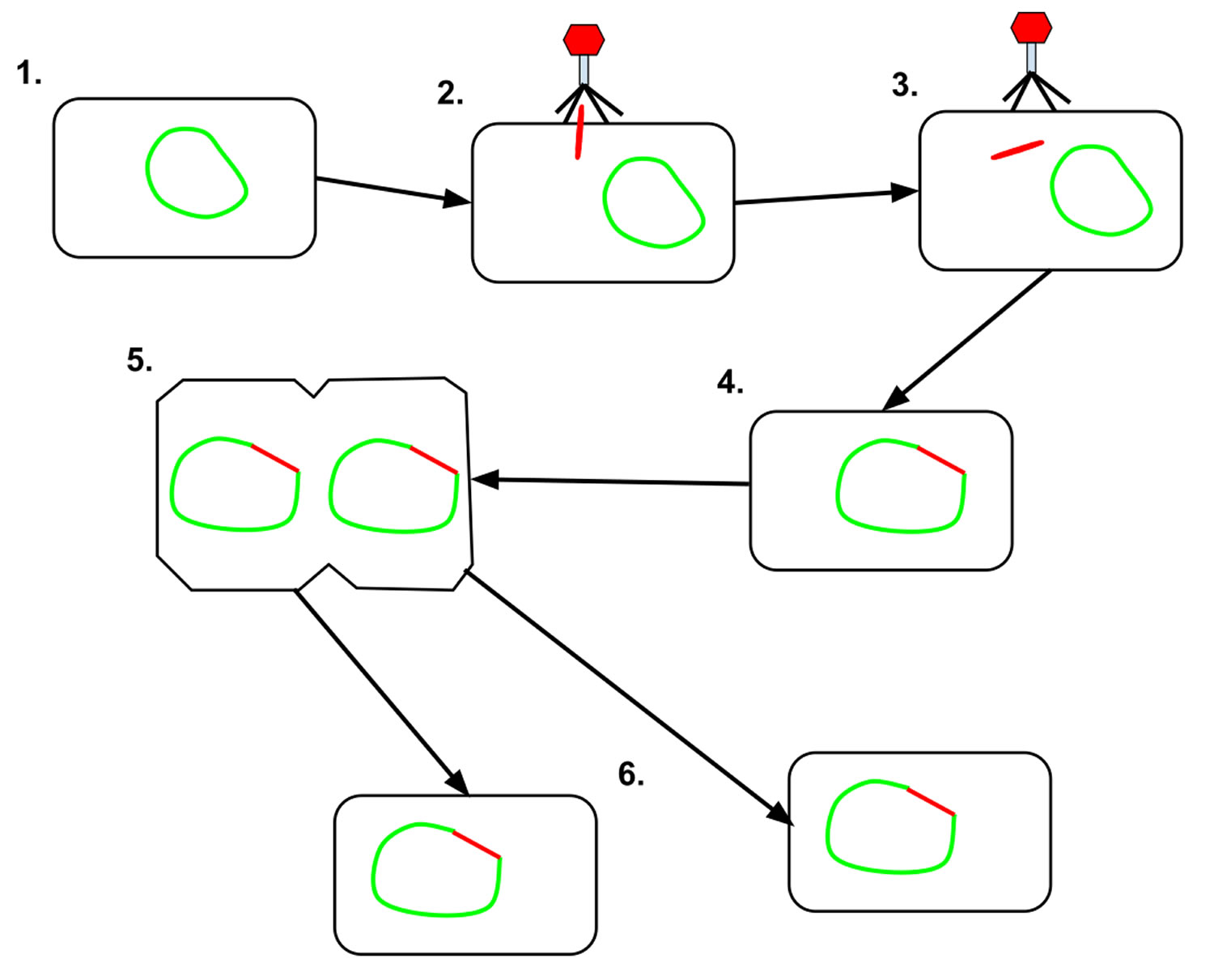
[Source 2 ]Figure 2. Lysogenic cycle
Footnote: 1. The prokaryotic cell is shown with its DNA, in green. 2. The bacteriophage attaches and releases its DNA, shown in red, into the prokaryotic cell. 3. The phage DNA then moves through the cell to the host’s DNA. 4. The bacteriophage DNA integrates itself into the host cell’s DNA, creating prophage. 5. The prophage then remains dominant until the host cell divides. 6. After the host cell has divided, the phage DNA in the daughter cells activate, and the phage DNA begins to express itself. Some of the cells containing the prophage go on to create new phages which will move on to infect other cells.
[Source 10 ]Difference between lytic and lysogenic cycle
In the lytic cycle, the phage redirects the host metabolisms towards the production of new phages, which are released during the lysis of the cell. In the lysogenic cycle, the genome of the phage typically remains in the host in a dormant stage (prophage) and replicates along with the host, until the lytic cycle is induced. A “lysogenic decision”, whether or not to establish a prophage state is made by the temperate phage after infection 2.
The difference between lysogenic and lytic cycles is that, in lysogenic cycles, the spread of the viral DNA occurs through the usual prokaryotic reproduction, whereas a lytic cycle is more immediate in that it results in many copies of the virus being created very quickly and the cell is destroyed. One key difference between the lytic cycle and the lysogenic cycle is that the lysogenic cycle does not lyse the host cell straight away 11. Phages that replicate only via the lytic cycle are known as virulent phages while phages that replicate using both lytic and lysogenic cycles are known as temperate phages 1.
In the lysogenic cycle, the phage DNA first integrates into the bacterial chromosome to produce the prophage. When the bacterium reproduces, the prophage is also copied and is present in each of the daughter cells. The daughter cells can continue to replicate with the prophage present or the prophage can exit the bacterial chromosome to initiate the lytic cycle.[1] In lysogenic cycle the host DNA is not hydrolysed but in lytic cycle the host DNA is hydrolysed in the lytic phase.
Lytic cycle of the phages
Lytic cycle of the phages consist of adsorption, separation of nucleic acids from protein coat, expression and replication of the nucleic acids, virion assembly and release. During the adsorption, the irreversible binding between a phage structure (e.g., tail fibers) and the receptor is accomplished. After adsorption, the cell wall is made penetrable, and the nucleic acid is transported into the cell, whereas the capsid remains outside the cell. Following injection, the genetic material stays in the cytoplasm. In this stage, gene expression, genome replication and morphogenesis occurs, i.e., the formation of the genomes and the capsids (and tails) and the packing of the genomes into the capsids. The phase of the latent period before capsids and genomes are assembled into mature phages is called the eclipse period. The rise period is characterized by the release of mature phages into the environment due to cell lysis and the detection of free phages (virions).
Lysogenic cycle of the phages
Lysogenic cycle of the phages consist of adsorption, separation of nucleic acids from protein coat, circularization of the phage chromosome, and repression of the phage genome. Following injection, the nucleic acid sequence is integrated into the host genome and, a phage coded protein, called a repressor, is made which binds to a particular site on the phage DNA, called the operator, and shuts off transcription of most phage genes except the repressor gene. However, when a lysogenic bacterium is exposed to adverse conditions, there is production of proteases which destroy the repressor protein. This in turn leads to the expression of the phage genes, reversal of the integration process and lytic multiplication.
Lysogenic conversion
In some interactions between lysogenic phages and bacteria, lysogenic conversion may occur, which can also be called phage conversion. It is when a temperate phage induces a change in the phenotype of the infected bacteria that is not part of a usual phage cycle. Changes can often involve the external membrane of the cell by making it impervious to other phages or even by increasing the pathogenic capability of the bacteria for a host. In this way, temperate bacteriophages also play a role in the spread of virulence factors, such as exotoxins and exoenzymes, amongst bacteria. This change then stays in the genome of the infected bacteria and is copied and passed down to daughter cells.
Bacterial survival
Lysogenic conversion has shown to enable biofilm formation in Bacillus anthracis 12 Strains of B. anthracis cured of all phage were unable to form biofilms, which are surface-adhered bacterial communities that enable bacteria to better access nutrients and survive environmental stresses 13. In addition to biofilm formation in B. anthracis, lysogenic conversion of Bacillus subtilis, Bacillus thuringiensis, and Bacillus cereus has shown an enhanced rate or extent of sporulation 12. Sporulation produces endospores, which are metabolically dormant forms of the bacteria that are highly resistant to temperature, ionizing radiation, desiccation, antibiotics, and disinfectants 12.
Bacterial virulence
Non-virulent bacteria have also been shown to transform into highly virulent pathogens through lysogenic conversion with the virulence factors carried on the lysogenic prophage 14. Virulence genes carried within prophages as discrete autonomous genetic elements, known as morons, confer an advantage to the bacteria that indirectly benefits the virus through enhanced lysogen survival 12.
Examples:
- Corynebacterium diphtheriae produces the toxin of diphtheria only when it is infected by the phage β. In this case, the gene that codes for the toxin is carried by the phage, not the bacterium 15.
- Vibrio cholerae is a non-toxic strain that can become toxic, producing cholera toxin, when it is infected with the phage CTXφ.
- Shigella dysenteriae, which produces dysentery has toxins that fall into two major groups, Stx1 and Stx2, whose genes are considered to be part of the genome of lambdoid prophages.
- Streptococcus pyogenes, produce a pyrogenic exotoxin, obtained by lysogenic conversion, which causes fever and a scarlet-red rash, scarlet fever.
- Certain strains of Clostridium botulinum, which causes botulism, express botulinum toxin from phage-tranduced genes.
Preventing lysogenic induction
Strategies to combat certain bacterial infections by blocking prophage induction (the transition from the lysogenic to the lytic cycle) by eliminating in vivo induction agents have been proposed 14. Reactive oxygen species (ROS), such as hydrogen peroxide, are strong oxidizing agents that can decompose into free radicals and cause DNA damage to bacteria, which leads to prophage induction 14. One potential strategy to combat prophage induction is through the use of glutathione, a strong antioxidant that can remove free radical intermediates 14. Another approach could be to cause an overexpression of CI repressor since prophage induction only occurs when the concentration of CI repressor is too low 14.
Clinical significance
Bacteriophages are clinically significant for several reasons. First, many highly pathogenic bacterial toxins are encoded by bacteriophage genomes, such that the host bacterium is only pathogenic when lysogenized by the toxin-encoding phage. Examples are cholera toxin in Vibrio cholerae 16, diphtheria toxin in Corynebacterium diphtheriae 17, botulinum neurotoxin in Clostridium botulinum 18, the binary toxin of Clostridium difficile 19, and Shiga toxin of Shigella species 20. Without their phage-encoded toxins, these bacterial species are either much less pathogenic or not pathogenic at all. Why phages encode these toxins is not known. While cholera toxin arguably helps both the phage and its host reach their next victim by inducing copious, watery diarrhea, the paralysis resulting from botulinum toxin would seem to have the opposite effect.
Second, bacteriophages are vectors for horizontal gene transfer, which may include antimicrobial resistance genes 21. They also have been engineered to introduce genes into specific strains for clinical effect, although this use is currently in the testing stage 22.
A third clinically relevant aspect of bacteriophages is that their detection can be used as a biomarker for the presence of their host in a complex environmental sample. This most commonly is used as a surrogate for fecal contamination of water sources. If the phage is present, the host most likely is as well. Alternatively, phages have been engineered to produce a detectable molecule, such as luciferase, when they infect their host as a means to detect bacteria in a mixed environmental sample 23.
While mostly supplanted by newer technologies, bacteriophages also are clinically relevant for their ability to distinguish strains of the same bacterial species. Most species of bacteria studied have multiple bacteriophage pathogens, just as humans as a species are susceptible to multiple viruses. Different strains within a species are resistant to some phages and not others. By infecting each strain systematically with a standardized panel of phages for that species, each strain can be identified by the pattern of susceptibility and resistance to each phage type. Phage typing of Staphylococcus aureus, for example, utilized a standardized panel of bacteriophages shared internationally to differentiate strains of Staphylococcus aureus. Before the development of molecular methods for this purpose, such as multilocus sequence typing and pulsed-field gel electrophoresis, phage typing was the criterion standard for tracking strains for epidemiological purposes 24.
Finally, bacteriophages were the first type of virus to be discovered and were a part of many of the fundamental discoveries of molecular biology. For example, the proof that DNA was the molecule that transmitted genetic information, the basic mechanisms of gene regulation, and the genetic code to name but a few, were all discovered using bacteriophages.
References- Campbell and Reece (2005). Biology. San Francisco: Pearson. pp. 338–339.
- Cruz-Tapias P, Castiblanco J, Correa NE, et al. Analysis of nucleic acids. In: Anaya JM, Shoenfeld Y, Rojas-Villarraga A, et al., editors. Autoimmunity: From Bench to Bedside [Internet]. Bogota (Colombia): El Rosario University Press; 2013 Jul 18. Chapter 46. Available from: https://www.ncbi.nlm.nih.gov/books/NBK459462
- Kasman LM, Porter LD. Bacteriophages. [Updated 2019 Aug 12]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2019 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK493185
- Ptashne M. Lambda’s switch: lessons from a module swap. Curr. Biol. 2006 Jun 20;16(12):R459-62.
- Chen; et al. (21 June 2005). “Population Fitness and the Regulation of Escherichia coli Genes by Bacterial Viruses”. PLOS Biology. 3 (7): e229. doi:10.1371/journal.pbio.0030229
- Stokar-Avihail A, Tal N, Erez Z, Lopatina A, Sorek R. Widespread Utilization of Peptide Communication in Phages Infecting Soil and Pathogenic Bacteria. Cell host & microbe. 2019 May 8;25(5):746-55
- Watson BNJ, Staals RHJ, Fineran PC. CRISPR-Cas-Mediated Phage Resistance Enhances Horizontal Gene Transfer by Transduction. MBio. 2018 Feb 13;9(1).
- De Sordi L, Lourenço M, Debarbieux L. The Battle Within: Interactions of Bacteriophages and Bacteria in the Gastrointestinal Tract. Cell Host Microbe. 2019 Feb 13;25(2):210-218.
- Maciejewska B, Olszak T, Drulis-Kawa Z. Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: an ambitious and also a realistic application? Appl. Microbiol. Biotechnol. 2018 Mar;102(6):2563-2581.
- Lysogenic cycle. https://en.wikipedia.org/wiki/Lysogenic_cycle
- Lodish; et al. (2008). Molecular Cell Biology. New York: W.H. Freeman. pp. 158–159.
- Louis-Charles Fortier; et al. (23 April 2013). “Importance of prophages to evolution and virulence of bacterial pathogens”. Virulence. 4 (5): 354–65. doi:10.4161/viru.24498
- Nadell; et al. (13 July 2011). “A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms”. PNAS. 108 (34): 14181–14185. Bibcode:2011PNAS..10814181N. doi:10.1073/pnas.1111147108
- Keen, Eric C. (14 December 2012). “Paradigms of pathogenesis: targeting the mobile genetic elements of disease”. Frontiers in Cellular and Infection Microbiology. 2: 161. doi:10.3389/fcimb.2012.00161
- Mokrousov I (January 2009). “Corynebacterium diphtheriae: genome diversity, population structure and genotyping perspectives”. Infection, Genetics and Evolution. 9 (1): 1–15. doi:10.1016/j.meegid.2008.09.011
- Pham TD, Nguyen TH, Iwashita H, Takemura T, Morita K, Yamashiro T. Comparative analyses of CTX prophage region of Vibrio cholerae seventh pandemic wave 1 strains isolated in Asia. Microbiol. Immunol. 2018 Oct;62(10):635-650.
- Holmes RK. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 2000 Feb;181 Suppl 1:S156-67.
- Fortier LC. The Contribution of Bacteriophages to the Biology and Virulence of Pathogenic Clostridia. Adv. Appl. Microbiol. 2017;101:169-200.
- Fortier LC. Bacteriophages Contribute to Shaping Clostridioides (Clostridium) difficile Species. Front Microbiol. 2018;9:2033.
- Doore SM, Schrad JR, Dean WF, Dover JA, Parent KN. Shigella Phages Isolated during a Dysentery Outbreak Reveal Uncommon Structures and Broad Species Diversity. J. Virol. 2018 Apr 15;92(8).
- Boyd EF. Bacteriophage-encoded bacterial virulence factors and phage-pathogenicity island interactions. Adv. Virus Res. 2012;82:91-118.
- Motlagh AM, Bhattacharjee AS, Goel R. Biofilm control with natural and genetically-modified phages. World J. Microbiol. Biotechnol. 2016 Apr;32(4):67.
- Schofield DA, Sharp NJ, Westwater C. Phage-based platforms for the clinical detection of human bacterial pathogens. Bacteriophage. 2012 Apr 01;2(2):105-283.
- Wiśniewska K, Szewczyk A, Piechowicz L, Bronk M, Samet A, Swieć K. The use of spa and phage typing for characterization of clinical isolates of methicillin-resistant Staphylococcus aureus in the University Clinical Center in Gdańsk, Poland. Folia Microbiol. (Praha). 2012 May;57(3):243-9.