What is RNA polymerase
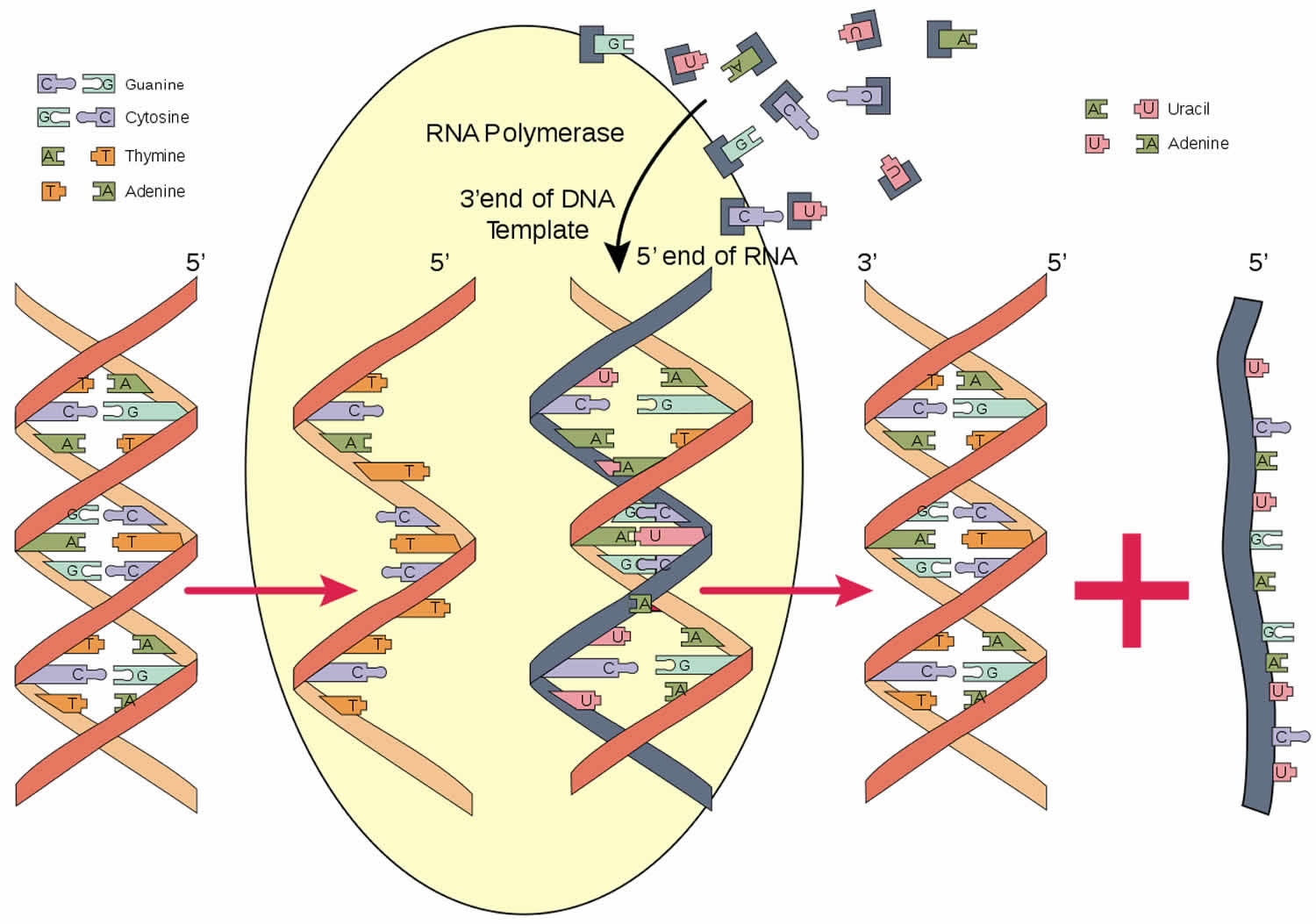
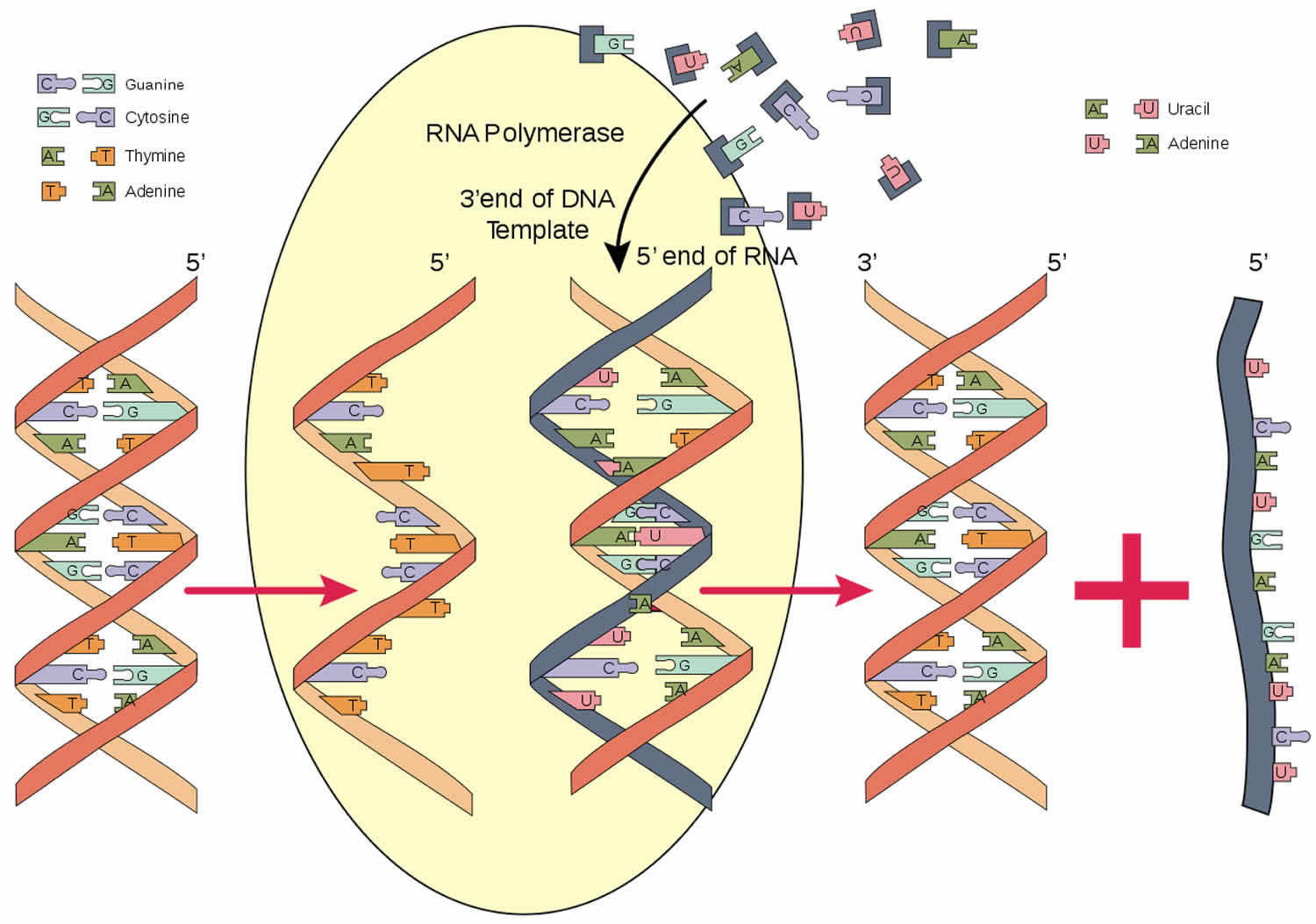
RNA polymerase is an enzyme that produces RNA (ribonucleic acid) and catalyzes the initiation and elongation of RNA chains from a DNA (deoxyribonucleic acid) template. RNA is created using a process known as transcription. The RNA polymerase is a key component to this process. The reaction that RNA polymerase enzyme catalyzes for is: (RNA)n + Ribonucleoside Triphosphate ->/<- (RNA)n+1 +PPi. RNA polymerases are relatively large. The size of RNA polymerase in a typical eukaryotic cell is roughly 500kDa. In bacteria it is roughly 400kDa and in T7 bacteriophage it is roughly 100kDa. Their speed of transcription is about 50 bases per second. A typical mRNA that codes for an average protein takes about 20 seconds in a prokaryotic cell and about 3 minutes in a eukaryotic cell. It is primarily longer in eukaryotes due to the fact that eukaryotic genes contain many segments that contain introns.
Figure 1. RNA polymerase function
What is the function of RNA polymerase?
RNA polymerase is an enzyme that is responsible for copying a DNA sequence into an RNA sequence, during the process of transcription. As complex molecule composed of protein subunits, RNA polymerase controls the process of transcription, during which the information stored in a molecule of DNA is copied into a new molecule of messenger RNA.
RNA polymerases have been found in all species, but the number and composition of these proteins vary across taxa. For instance, bacteria contain a single type of RNA polymerase, while eukaryotes (multicellular organisms and yeasts) contain three distinct types. In spite of these differences, there are striking similarities among transcriptional mechanisms. For example, all species require a mechanism by which transcription can be regulated in order to achieve spatial and temporal changes in gene expression.
Gene transcription takes place in the nucleus of eukaryotic cells and transcription is performed by three different multisubunit RNA polymerases, RNA polymerase I, RNA polymerase II, and RNA polymerase III. Still little is known today about the biogenesis of these RNA polymerases: from their origin of synthesis, the cytoplasm, to their arrival in the nucleus for transcription. Only until recently have studies shown that polymerase assembly intermediates, assembly factors and factors required for polymerase nuclear import exist in the cell cytoplasm. RNA polymerase II is the most identifiable one so is the basis of most studies on the biogenesis of RNA polymerase.
RNA polymerase II transcribes mRNAs and small non-coding RNAs and contains 12 polypeptide subunits. Each RNA polymerase has their own specified role in RNA polymerase. They all have ten identical subunit catalytic cores. The peripheral subunits are what differentiate their structure and function; RNA polymerase II has been determined to contain cores that allow it to model the homologous cores in RNA polymerase I and RNA polymerase III. RNA polymerase I and RNA polymerase III will bind to opposite sides of RNA polymerase II (binding to Rpb1 and Rpb2) and are then divided into three interacting subunits.
The assembly of eukaryotic RNA core was first identified in studies of bacterial RNA polymerase because RNA polymerase II core subunits are exactly identical to that of bacteria. Assembly of RNA polymerase II is initiated by the formation of the αα dimer which interacts with the β and forms a bound complex intermediate. The active cleft in the RNA polymerase II is composed of β subunits which are formed in the final step of assembly, so the polymerase will not be catalytically active until it is complete. RNA polymerase II in both bacteria and eukaryotic cells has both exhibited formation in equivalent manner.
Assembly in vitro experiments have also been conducted to determine the origins of RNA polymerase II. Using three mutant large subunits, their assembly was followed with the use of pulse chase experiments. Scientists found that Rpb3 and Rpb3 were the first to interact, and the bound complex then interacts with Rpb1. However, because larger mutated subunits were used, final assembly could not be complete without the use of Rpb6, Rpb10, and Rpb12, which are not normally part of final assembly in normal sized RNA polymerase II.
If any RNA subunits are lost during its assembly, there will be an excess of Rpb1 present in the cytoplasm, meaning that the polymerase needs to be fully assembled before it is allowed to enter the nucleus and take place in transcriptase. RNA polymerase II nuclear localization factors have been identified to be functional polymerase-interacting proteins in the cell. The accumulation of Rpb1 is caused by the depletion of GPN1 and GPN3. The expression of GPN1 will lead to the depletion of excess Rpb1. GPN1 binding to RNA polymerase II can also be directly influence the ability of GTP to bind properly. Homologs of GPN1 also aid in the biogenesis and final assembly of polymerase II. GPN1 interacts with the CCT complex, which chaperones many subunits in the formation of RNA polymerase II.
DNA polymerase vs RNA polymerase
DNA polymerase is a type of enzyme that is responsible for forming new copies of DNA, in the form of nucleic acid molecules. Nucleic acids are polymers, which are large molecules made up of smaller, repeating units that are chemically connected to one another. DNA is composed of repeating units called nucelotides or nucleotide bases. DNA polymerase is responsible for the process of DNA replication, during which a double-stranded DNA molecule is copied into two identical DNA molecules. Scientists have taken advantage of the power of DNA polymerase molecules to copy DNA molecules in test tubes via polymerase chain reaction, also known as PCR.
The synthesis of RNA and DNA is similar in many aspects. Both of them follow the synthesis direction of 5′->3′. Another is that the method of elongation is by the 3’OH group at the terminus of the growing chain that makes a nucleophilic attack on the innermost phosphate of the incoming nucleoside triphosphate. Another similarity is that the synthesis is driven by the hydrolysis of pyrophosphate. However the difference between the two is that RNA polymerase does not require a primer unlike DNA polymerase which does. Also although DNA polymerase can actually correct mistakes in the nucleotide transcription, RNA polymerase lacks this ability to excise the mismatches nucleotides.