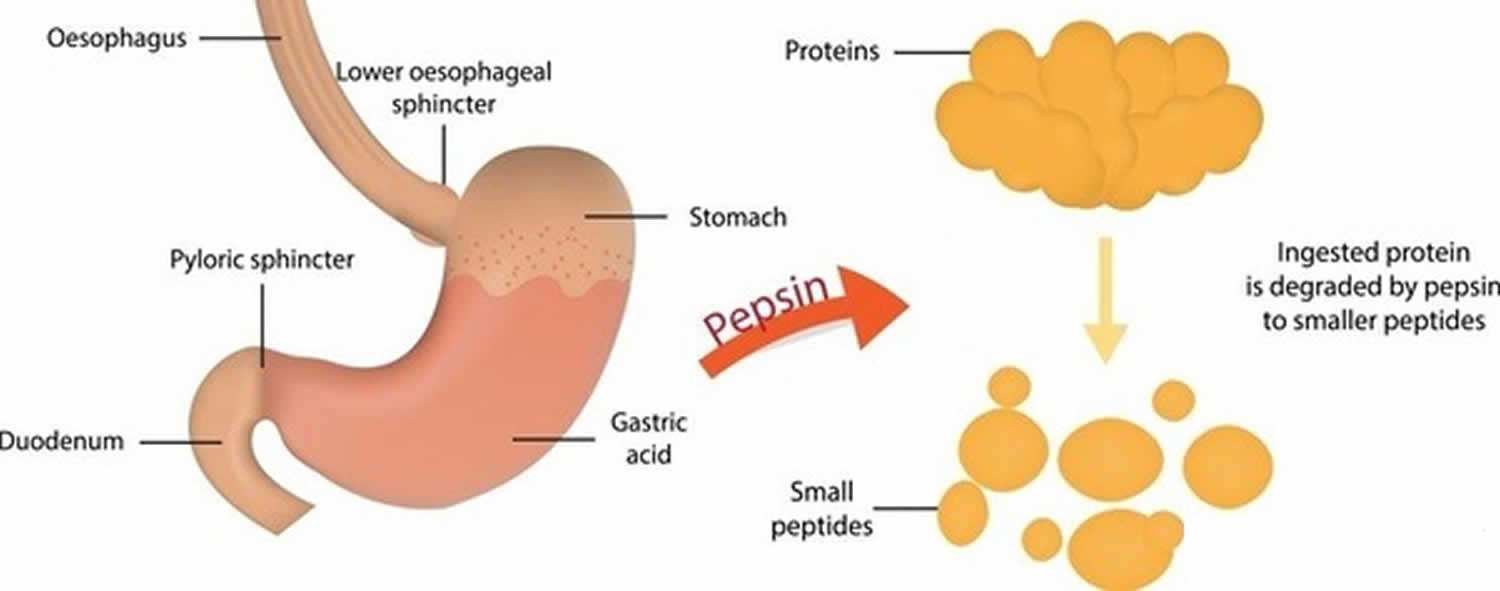
What is pepsin
Pepsin is an aspartic acid protease enzyme that uses aspartic acid residues in the active center 1 to activate a water molecule and use it to cleave protein chains. Pepsin a protease enzyme breaks down proteins into smaller peptides. Chief cells in the stomach secrete pepsin to help you digest or breaks down proteins into smaller compounds called peptides. In fact, pepsin is only produced in your stomach. If pepsin is found in other parts of the body, e.g., in saliva or sputum samples, the only mechanism by which that would be possible is via reflux of gastric contents. Pepsin has been identified within the larynx (voicebox) in the saliva of patients with suspected reflux laryngitis and in the exhaled breath of those with airways and lung disease, where reflux is thought to have played a part. At the mean pH of the laryngopharynx (pH = 6.8) pepsin would be inactive but could be reactivated upon subsequent acid reflux events resulting in damage to local tissues. Pepsin has also been found in the middle ear in otitis media effusions (glue ear). Pepsin is active at the acidity of normal gastric content – pH1.0 to pH 4.5, but is easily denatured by pH above 7.0 2.
Pepsin is produced in your stomach by the Chief cells and is one of the main digestive enzymes in the digestive systems of humans and many other animals, where it helps digest the proteins in food. Pepsin works its best in strong hydrochloric acid.
Pepsin are stored as inactive zymogens in granules of the chief cells of human gastric mucosa 3. There are five types of zymogens of pepsins, gastric digestive proteinases, are known: pepsinogens A, B, and F, progastricsin, and prochymosin 4. The amino acid and/or nucleotide sequences of more than 50 pepsinogens other than pepsinogen B have been determined to date 5. Phylogenetic analyses based on these sequences indicate that progastricsin diverged first followed by prochymosin, and that pepsinogens A and F are most closely related.
Pepsin has 330–350 amino acid residues with mainly β-sheets. Pepsin has a three-dimensional structure, of which one or more polypeptide chains twist and fold, bringing together a small number of amino acids to form the active site, or the location on the enzyme where the substrate binds and the reaction takes place. Tertiary structures, clarified by X-ray crystallography, are commonly bilobal with a large active-site cleft between the lobes. The catalytic site of pepsin is located in the middle narrow slit of two lobes; two aspartic acid residues, Asp32 and Asp215, are located at the end of each domain 6. These two aspartic acid residues are connected through a low-barrier hydrogen bond. Findings from recent decades suggested that this low barrier hydrogen bond is the key feature in the catalytic mechanism for aspartic proteases, which facilitates proton transfer during catalysis 7.
Two aspartates in the center of the cleft, Asp32 and Asp215, function as catalytic residues, and thus pepsinogens are classified as aspartic proteinases. Conversion of pepsinogens (inactive form of pepsin) to pepsins proceeds autocatalytically at acidic pH (pH <2.0) by two different pathways, a one-step pathway to release the intact activation segment directly, and a stepwise pathway through a pseudo-pepsin(s). The active-site cleft is large enough to accommodate at least seven residues of a substrate, thus forming S4 through S’3 subsites. Hydrophobic and aromatic amino acids are preferred at the P1 and P’1 positions. Interactions at additional subsites are important in some cases, for example with cleavage of kappa-casein by chymosin.
Figure 1. Pepsin chemical structure (bilobal structure of pepsin)
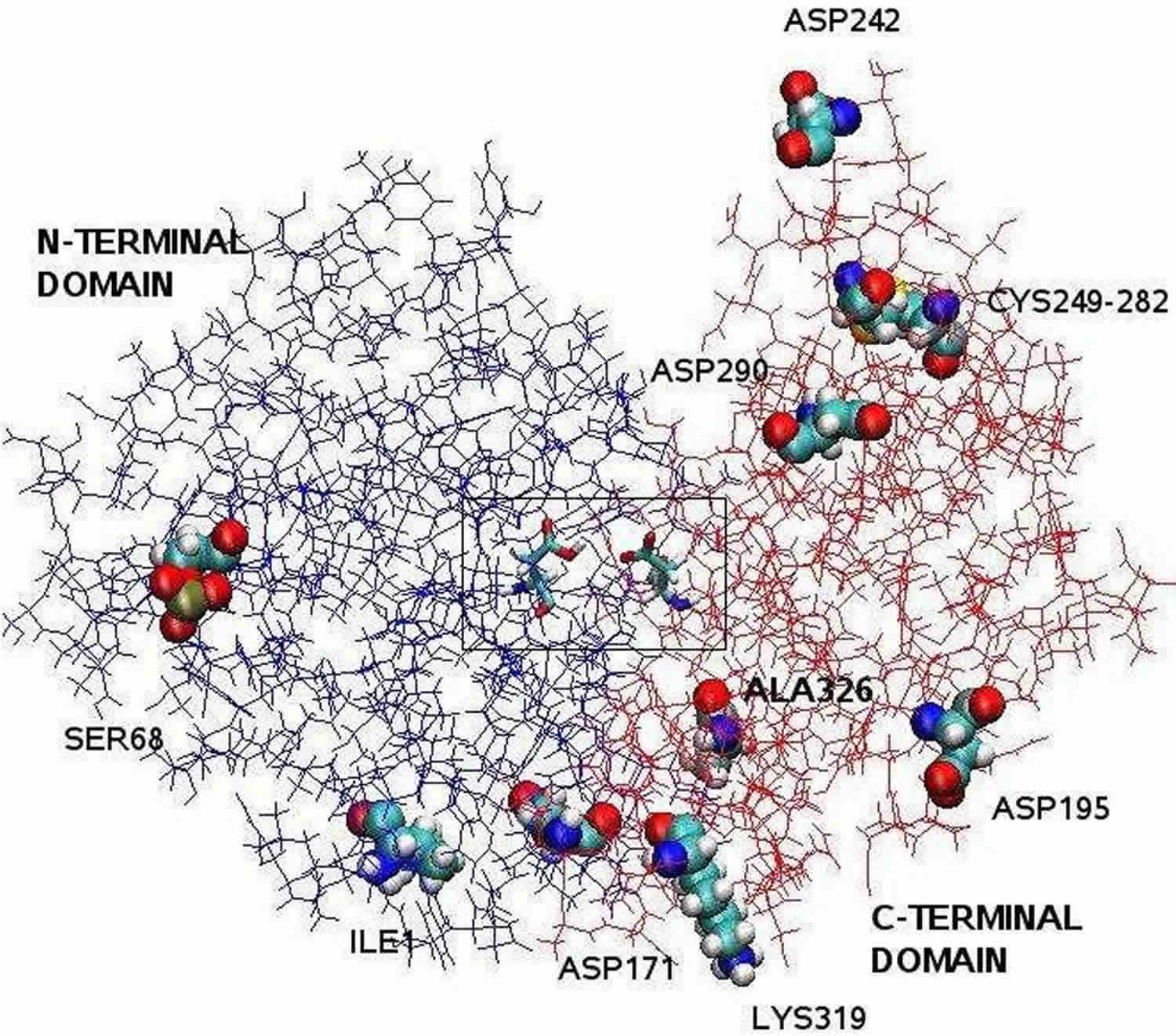 Footnote: The pepsin structure clarified by X-ray crystallography is bilobal with a large active site cleft between the two lobes. Residues 1 to 175 form the N-terminal domain, whereas residues 176 to 326 constitute the C-terminal domain. Two aspartates in the center of the cleft, Asp32 and Asp215, function as catalytìc residues. The catalytic site, consisting of Asp32 and Asp215, is located in the center of the cleft. The charged amino acid residues located on the protein surface are, Asp171, Asp195, Asp242, Asp290, Lys319, Ile1 (N-terminal one), Ala326 (C-terminal one). The phosphorylated Ser68 is fully exposed to the solvent and the disulphide bridge between Cys 249 and Cys 282 is located on the protein surface.
[Source 8 ]
Footnote: The pepsin structure clarified by X-ray crystallography is bilobal with a large active site cleft between the two lobes. Residues 1 to 175 form the N-terminal domain, whereas residues 176 to 326 constitute the C-terminal domain. Two aspartates in the center of the cleft, Asp32 and Asp215, function as catalytìc residues. The catalytic site, consisting of Asp32 and Asp215, is located in the center of the cleft. The charged amino acid residues located on the protein surface are, Asp171, Asp195, Asp242, Asp290, Lys319, Ile1 (N-terminal one), Ala326 (C-terminal one). The phosphorylated Ser68 is fully exposed to the solvent and the disulphide bridge between Cys 249 and Cys 282 is located on the protein surface.
[Source 8 ]Pepsin, as an aspartic protease, has a very low pH optimum. Kondjoyan, Daudin, and Santé-Lhoutellier 9 studied the digestibility of myofibrillar proteins found maximum pepsin activity at pH around 2. Pletschke, Naudé, and Oelofsen 10 studied the hydrolysis of hemoglobin by porcine and ostrich pepsin, and also found an optimum pH of 2.0 for pepsin activity. Once activated by pH <4.5, pepsin enzyme are easily denatured by pH above 7.0, constituting properties that determine the characteristics of pepsin 2.
In contrast to the influence of pH, the effect of ionic strength on enzyme activity is less often discussed. Previous work by Dale and White 11 showed that increasing the ionic strength clearly decreases the reaction rate of immobilized pancreatic ribonuclease, while Butré, Wierenga, and Gruppen 12 studied the enzymatic hydrolysis of whey protein isolate by alcalase and neutrase, and also found that the presence of 0.5M NaCl decreased the rate of hydrolysis.
Two potent naturally occurring inhibitors of pepsin are known: pepstatin, a pentapeptide from Streptomyces, and a unique proteinous inhibitor from Ascaris. Pepsin also undergoes feedback inhibition; a product of protein digestion slows down the reaction by inhibiting pepsin 13.
Where is pepsin produced?
The Chief cells secrete pepsin in the form of an inactive enzyme precursor called pepsinogen. When pepsinogen contacts hydrochloric acid from the parietal cells, it breaks down rapidly, forming pepsin 14. Pepsin begins the digestion of nearly all types of dietary protein into polypeptides. This enzyme is most active in an acidic environment, which is provided by the hydrochloric acid in gastric juice.
Figure 2. Stomach cells (gastric glands)
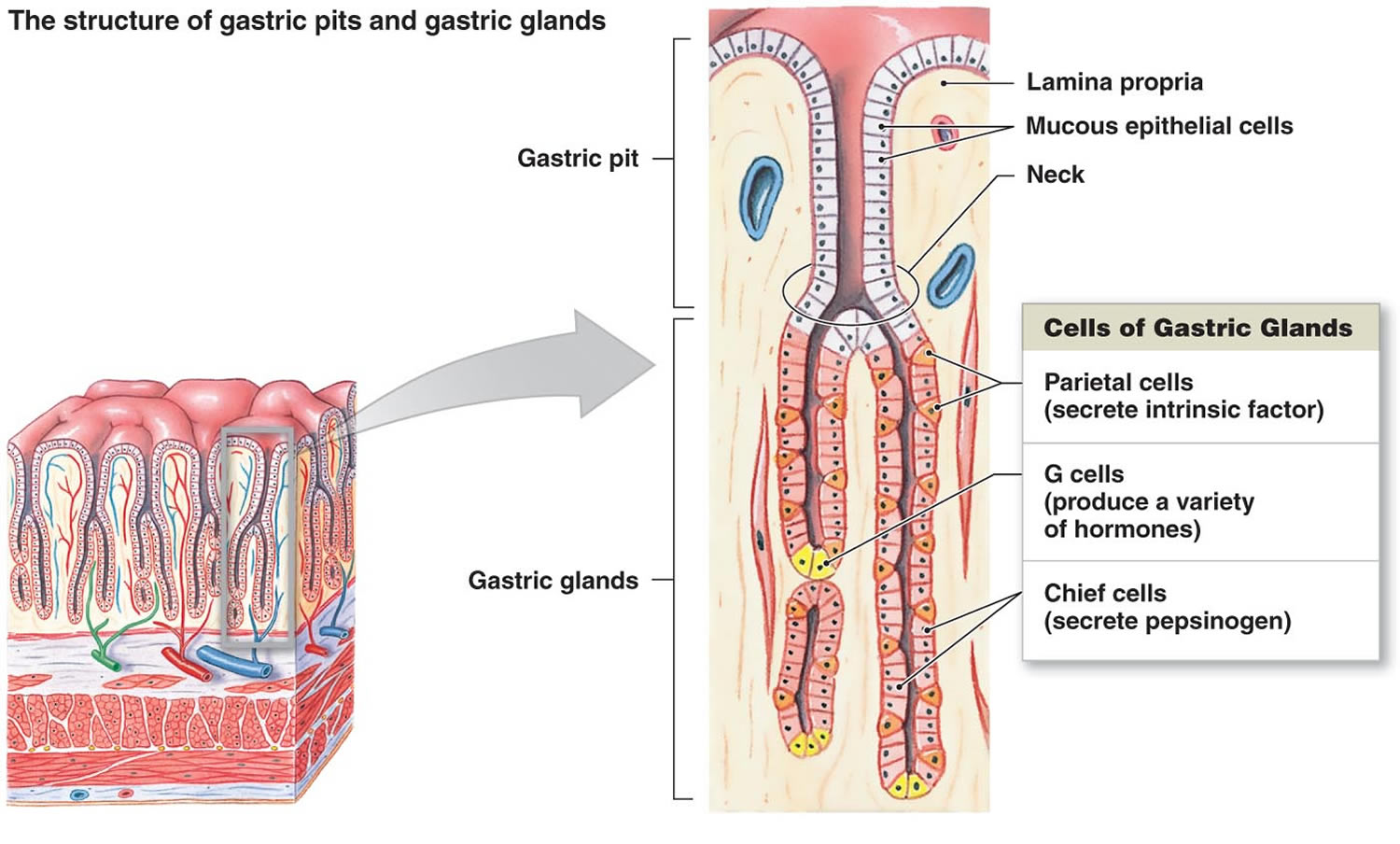 Footnote: Lining of the stomach. Gastric glands include mucous cells, parietal cells, and chief cells. The mucosa of the stomach is studded with gastric pits that are the openings of the gastric glands.
Footnote: Lining of the stomach. Gastric glands include mucous cells, parietal cells, and chief cells. The mucosa of the stomach is studded with gastric pits that are the openings of the gastric glands.
What does pepsin do?
Pepsin formed from pepsinogen (inactive form of pepsin) in the presence of hydrochloric acid, digests nearly all types of dietary protein into polypeptides (smaller protein compounds). Pepsin digests proteins in strong stomach acid. Pepsin is optimized for digestion of food in the acidic environment of the stomach and is the first protease that food proteins encounter in the digestive tract. Pepsin is most active in acidic environments between 37 °C and 42 °C 15.
The specificity of pepsin (i.e. the cleavage probability of peptide bonds by pepsin) is mainly influenced by the amino acid residues at position P1 and P1′, while the amino acid residues at other subsites may also play a role. In general, pepsin prefers to cleave after phenylalanine, leucine, and methionine, whereas it rarely cleaves after histidine, lysine, proline, and arginine 5. Aromatic residues tyrosine, tryptophan, and phenylalanine are favored at position P1′. Proline is strongly disfavored at P2, P2′, and P3′ position, while histidine, lysine, and arginine are disfavored at the P3 position. The P4 and P4′ positions have little influence 16.
Figure 3. Pepsin function
- Cooper JB. Aspartic proteinases in disease: a structural perspective. Curr Drug Targets 2002; 3: 155–173.
- Taylor WH. Biochemistry of the pepsins. In: Code CF, Heide W, eds. Handbook of Physiology‐Alimentary Canal, vol V. Baltimore: Williams & Wilkins, 1968: 2567–2587.
- Kageyama T. Pepsinogens, progastricsins and prochymosins: structure, function, evolution and development. Cell Mol Life Sci 2002; 59: 288–306.
- Pepsinogens, progastricsins, and prochymosins: structure, function, evolution, and development. Cell Mol Life Sci. 2002 Feb;59(2):288-306. https://www.ncbi.nlm.nih.gov/pubmed/11915945
- Revisiting the enzymatic kinetics of pepsin using isothermal titration calorimetry. Food Chemistry 268 (2018) 94–100 https://doi.org/10.1016/j.foodchem.2018.06.042
- Dunn, B.M. (2001). Overview of pepsin-like aspartic peptidases. Current protocols in protein science, Chapter 21, Unit 21.3. DOI: 10.1002/0471140864.ps2103s25
- Northrop, D. B. (2001). Follow the protons: A Low-barrier hydrogen bond unifies the mechanisms of the aspartic proteases. Accounts of Chemical Research, 34, 790–797. http://dx.doi.org/10.1021/ar000184m
- Giussani, L & Fois, Ettore & Gianotti, Enrica & Tabacchi, Gloria & Gamba, A & Coluccia, Salvatore. (2008). The determination of pepsin dimensions at different p H values: A simulation study. Il Nuovo Cimento B. 123. 1477-1483. 10.1393/ncb/i2008-10723-3.
- Kondjoyan, A., Daudin, J.-D., & Santé-Lhoutellier, V. (2015). Modelling of pepsin digestibility of myofibrillar proteins and of variations due to heating. Food Chemistry, 172, 265–271. http://dx.doi.org/10.1016/j.foodchem.2014.08.110
- Pletschke, B. I., Naudé, R. J., & Oelofsen, W. (1995). Ostrich pepsins I and II: A kinetic and thermodynamic investigation. The International Journal of Biochemistry & Cell Biology, 27, 1293–1302. http://dx.doi.org/10.1016/1357-2725(95)00092-4
- Dale, B. E., & White, D. H. (1983). Ionic strength: A neglected variable in enzyme technology. Enzyme and Microbial Technology, 5, 227–229. http://dx.doi.org/10.1016/0141-0229(83)90101-1
- Butré, C. I., Wierenga, P. A., & Gruppen, H. (2012). Effects of ionic strength on the enzymatic hydrolysis of diluted and concentrated whey protein isolate. Journal of Agricultural and Food Chemistry, 60, 5644–5651. http://dx.doi.org/10.1021/jf301409n
- Greenwell P, Knowles JR, Sharp H (Jun 1969). “The inhibition of pepsin-catalysed reactions by products and product analogues. Kinetic evidence for ordered release of products”. The Biochemical Journal. 113 (2): 363–8 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1184643/pdf/biochemj00700-0137.pdf
- Kageyama, T. (2014). Pepsinogens, progastricsins, and prochymosins: Structure, function, evolution, and development. Cellular and Molecular Life Sciences CMLS, 59, 288–306. http://dx.doi.org/10.1007/s00018-002-8423-9
- Information on EC 3.4.23.1 – pepsin A. https://www.brenda-enzymes.org/enzyme.php?ecno=3.4.23.1#pH%20OPTIMUM
- Ahn, J., Cao, M.-J., Yu, Y.Q., & Engen, J.R. (2013). Accessing the reproducibility and specificity of pepsin and other aspartic proteases. Biochimica et Biophysica Acta (BBA) – Proteins and Proteomics, 1834, pp. 1222–1229, DOI: 10.1016/j.bbapap.2012.10.003